Computing mass and charge density on each axis
Here we compute the mass and charge density of water along the three cartesian axes of a fixed-volume unit cell (i.e. from a simulation in the NVT ensemble).
Last updated: December 2022 with MDAnalysis 2.4.0-dev0
Minimum version of MDAnalysis: 0.17.0
Packages required:
[1]:
import MDAnalysis as mda
from MDAnalysis.tests.datafiles import waterPSF, waterDCD
from MDAnalysis.analysis import lineardensity as lin
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
Loading files
The test files we are working with are a cube of water.
[2]:
u = mda.Universe(waterPSF, waterDCD)
/Users/lily/micromamba/envs/mda-user-guide-dev/lib/python3.10/site-packages/MDAnalysis/coordinates/DCD.py:165: DeprecationWarning: DCDReader currently makes independent timesteps by copying self.ts while other readers update self.ts inplace. This behavior will be changed in 3.0 to be the same as other readers. Read more at https://github.com/MDAnalysis/mdanalysis/issues/3889 to learn if this change in behavior might affect you.
warnings.warn("DCDReader currently makes independent timesteps"
MDAnalysis.analysis.lineardensity.LinearDensity (API docs) will partition each of your axes into bins of user-specified binsize (in angstrom), and give the average mass density and average charge density of your atom group selection.
This analysis is only suitable for a trajectory with a fixed box size. While passing a trajectory with a variable box size will not raise an error, LinearDensity will not account for changing dimensions. It will only evaluate the density of your atoms in the bins created from the trajectory frame when the class is first initialised.
Below, we iterate through the trajectory to verify that its box dimensions remain constant.
[3]:
for ts in u.trajectory:
print(ts.dimensions)
[50. 50. 50. 90. 90. 90.]
[50. 50. 50. 90. 90. 90.]
[50. 50. 50. 90. 90. 90.]
[50. 50. 50. 90. 90. 90.]
[50. 50. 50. 90. 90. 90.]
[50. 50. 50. 90. 90. 90.]
[50. 50. 50. 90. 90. 90.]
[50. 50. 50. 90. 90. 90.]
[50. 50. 50. 90. 90. 90.]
[50. 50. 50. 90. 90. 90.]
You can choose to compute the density of individual atoms, residues, segments, or fragments (groups of bonded atoms with no bonds to any atom outside the group). By default, the grouping is for atoms.
[4]:
density = lin.LinearDensity(u.atoms,
grouping='atoms').run()
The results of the analysis are in density.results.
[5]:
density.nbins
[5]:
np.int64(200)
[6]:
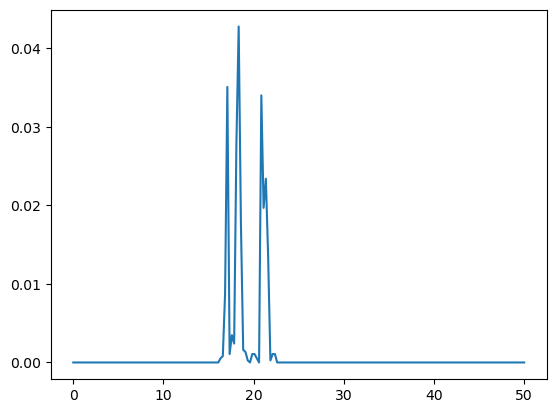
density.results['x']['mass_density']
[6]:
array([0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0.00053562, 0.00080344, 0.00876945, 0.03507781, 0.00107125,
0.00348155, 0.00241031, 0.02791523, 0.04277601, 0.0175389 ,
0.00160687, 0.00133906, 0.00026781, 0. , 0.00107125,
0.00107125, 0.00053562, 0. , 0.03400656, 0.0196814 ,
0.02339659, 0.0135559 , 0.00026781, 0.00107125, 0.00107125,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ])
[7]:
density.results['x'].keys()
[7]:
KeysView({'dim': 0, 'slice_volume': np.float64(625.0), 'mass_density': array([0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0.00053562, 0.00080344, 0.00876945, 0.03507781, 0.00107125,
0.00348155, 0.00241031, 0.02791523, 0.04277601, 0.0175389 ,
0.00160687, 0.00133906, 0.00026781, 0. , 0.00107125,
0.00107125, 0.00053562, 0. , 0.03400656, 0.0196814 ,
0.02339659, 0.0135559 , 0.00026781, 0.00107125, 0.00107125,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ]), 'mass_density_stddev': array([0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0.00107125, 0.00122727, 0.01688797, 0.01691979, 0.00177646,
0.00241031, 0.00279604, 0.02179554, 0.02689655, 0.02096112,
0.001312 , 0.00133906, 0.00080344, 0. , 0.001312 ,
0.001312 , 0.00107125, 0. , 0.01700328, 0.03402765,
0.02131476, 0.01957657, 0.00080344, 0.001312 , 0.001312 ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ]), 'charge_density': array([ 0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0.00022158, 0.00033237, -0.00033237, -0.00132949, 0.00044316,
0.00144029, 0.00099712, -0.00033237, -0.00210503, -0.00066475,
0.00066475, 0.00055396, 0.00011079, 0. , 0.00044316,
0.00044316, 0.00022158, 0. , -0.00177266, 0.00022158,
-0.00022158, -0.00033237, 0.00011079, 0.00044316, 0.00044316,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ]), 'charge_density_stddev': array([0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0.00044316, 0.00050771, 0.00099712, 0.00108553, 0.00073491,
0.00099712, 0.00115669, 0.00111344, 0.00144029, 0.00112985,
0.00054276, 0.00055396, 0.00033237, 0. , 0.00054276,
0.00054276, 0.00044316, 0. , 0.00088633, 0.0018406 ,
0.00129204, 0.00111344, 0.00033237, 0.00054276, 0.00054276,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0. , 0. , 0. , 0. , 0. ,
0.
[8]:
density.results['y']['dim']
[8]:
1
[9]:
plt.plot(np.linspace(0, 50, 200), density.results['x']['mass_density'])
[9]:
[<matplotlib.lines.Line2D at 0x156168ee0>]
References
[1] Richard J. Gowers, Max Linke, Jonathan Barnoud, Tyler J. E. Reddy, Manuel N. Melo, Sean L. Seyler, Jan Domański, David L. Dotson, Sébastien Buchoux, Ian M. Kenney, and Oliver Beckstein. MDAnalysis: A Python Package for the Rapid Analysis of Molecular Dynamics Simulations. Proceedings of the 15th Python in Science Conference, pages 98–105, 2016.
URL: https://conference.scipy.org/proceedings/scipy2016/oliver_beckstein.html, doi:10.25080/Majora-629e541a-00e.
[2] Naveen Michaud-Agrawal, Elizabeth J. Denning, Thomas B. Woolf, and Oliver Beckstein. MDAnalysis: A toolkit for the analysis of molecular dynamics simulations. Journal of Computational Chemistry, 32(10):2319–2327, July 2011.
URL: http://doi.wiley.com/10.1002/jcc.21787, doi:10.1002/jcc.21787.